STL plugin for Orthanc¶
Contents
Overview¶
This official plugin by the ICTEAM institute of UCLouvain extends Orthanc with support for Encapsulated 3D Manufacturing Model IODs. As of release 1.0 of the plugin, this support is limited to STL files.
The plugin allows to attach STL files to existing DICOM studies and to generate a STL mesh from structure sets (i.e., DICOM RT-STRUCT) or from NIfTI binary volumes. A high-level description of these features is available as a paper.
Importantly, any creation of a STL file requires the version of Orthanc to be above or equal to 1.12.1.
For researchers: Please cite this paper.
Compilation¶
Official releases can be downloaded from the Orthanc homepage. As an alternative, the repository containing the source code can be accessed using Mercurial.
The procedure to compile this plugin is similar of that for the core of Orthanc. The following commands should work on most GNU/Linux distributions, provided Docker is installed:
$ mkdir Build
$ cd Build
$ ../Resources/CreateJavaScriptLibraries.sh
$ cmake .. -DSTATIC_BUILD=ON -DCMAKE_BUILD_TYPE=Release
$ make
The compilation will produce a shared library libOrthancSTL.so
that contains the STL plugin for Orthanc.
Pre-compiled Linux Standard Base (LSB) binaries are available for download.
Pre-compiled binaries for Microsoft Windows and macOS are available as well.
Furthermore, the Docker images
jodogne/orthanc-plugins and orthancteam/orthanc also contain the
plugin. Debian and Ubuntu packages can be found in the
standalone repository
https://debian.orthanc-labs.com/.
Usage using Orthanc Explorer¶
The plugin extends the default Orthanc Explorer Web interface with some new features.
Attach STL file¶
An existing STL file can be attached to an existing DICOM study by clicking on the “Attach STL model” yellow button:
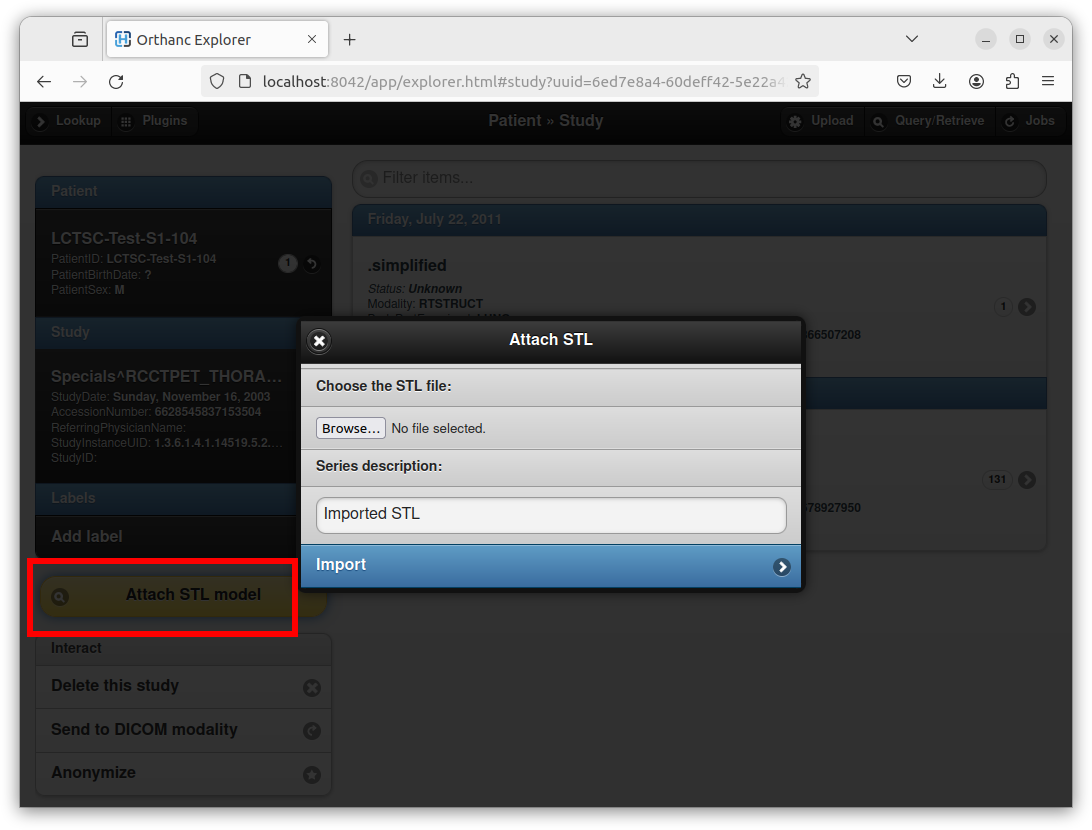
After selecting the STL file, entering a series description, and clicking on the “Import” button, Orthanc creates a new DICOM instance that embeds the STL file. Orthanc Explorer then automatically opens the parent DICOM series containing the newly created DICOM STL instance. A button entitled “STL viewer” can then be used to render the STL:
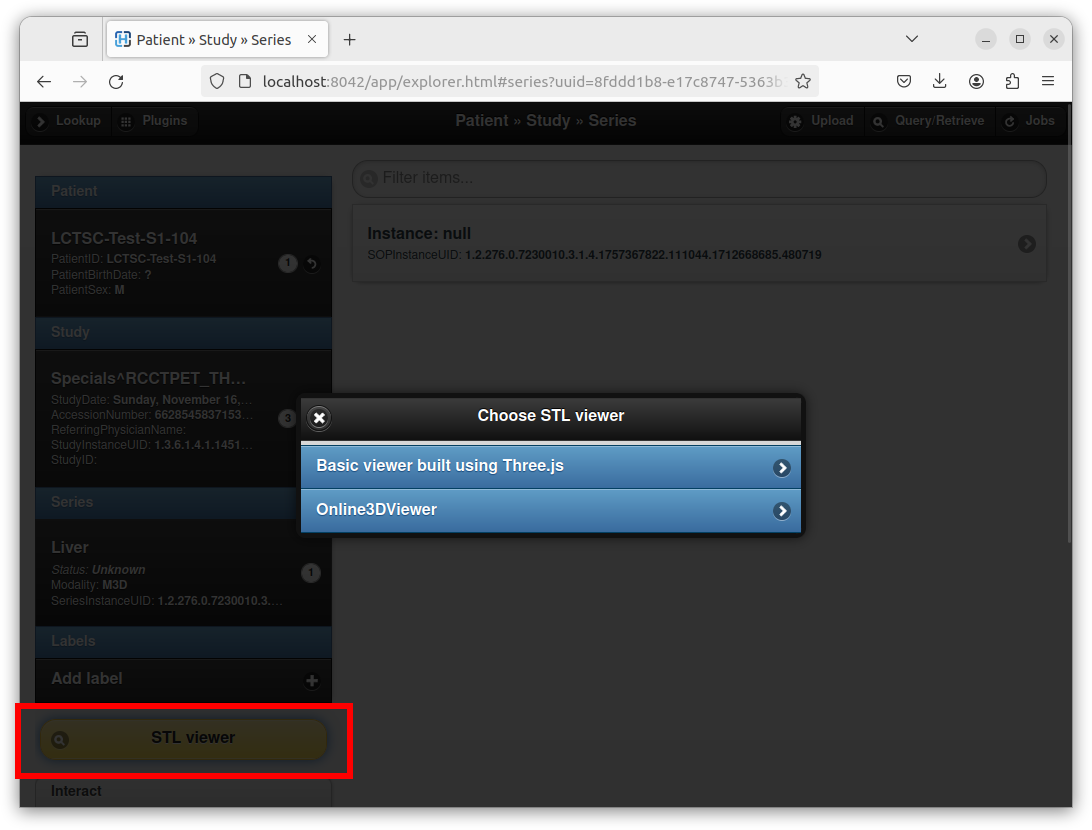
Note how the STL plugin provides two viewers:
One very basic custom viewer with a small footprint that is directly built using the well-known Three.js library.
One slightly more advanced Web viewer that corresponds to Online 3D Viewer running in engine mode.
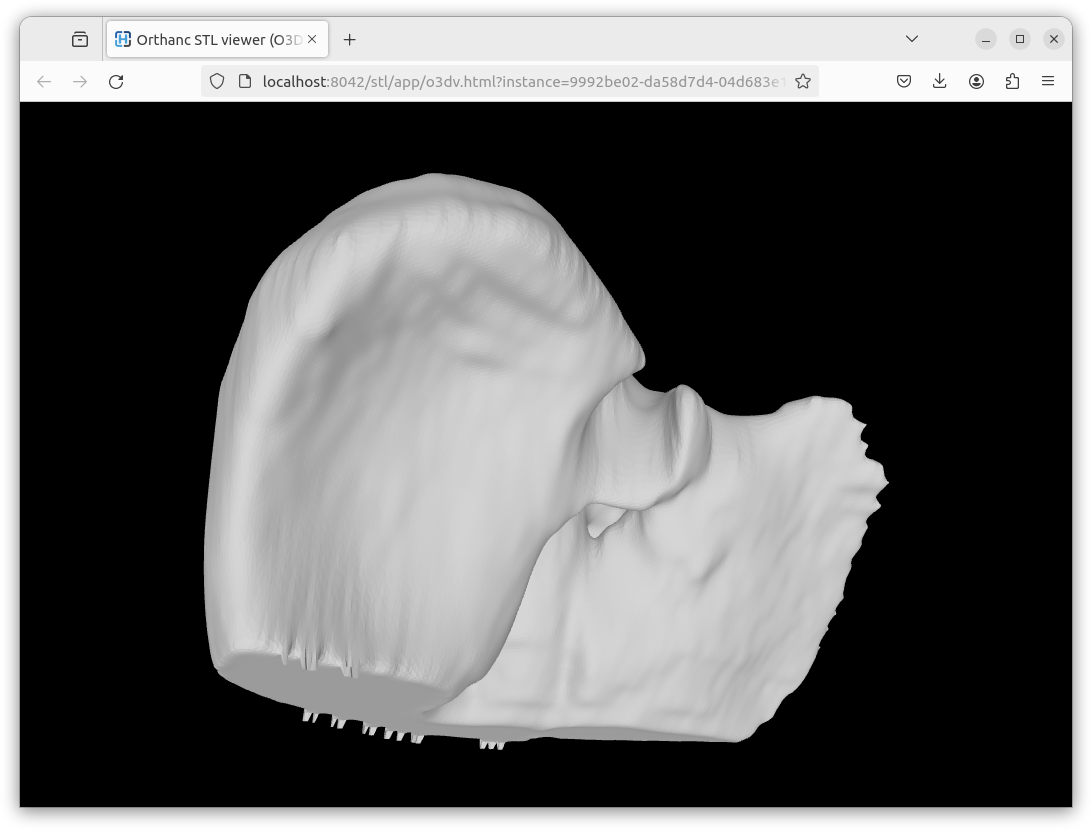
Here is a screenshot of a rendering using the Online3DViewer embedded viewer:
Create STL from RT-STRUCT¶
The plugin can also be used to create a STL model from DICOM structure sets (RT-STRUCT) that are routinely used in the context of radiotherapy and nuclear medicine. To this end, open a DICOM RT-STRUCT series using Orthanc Explorer:
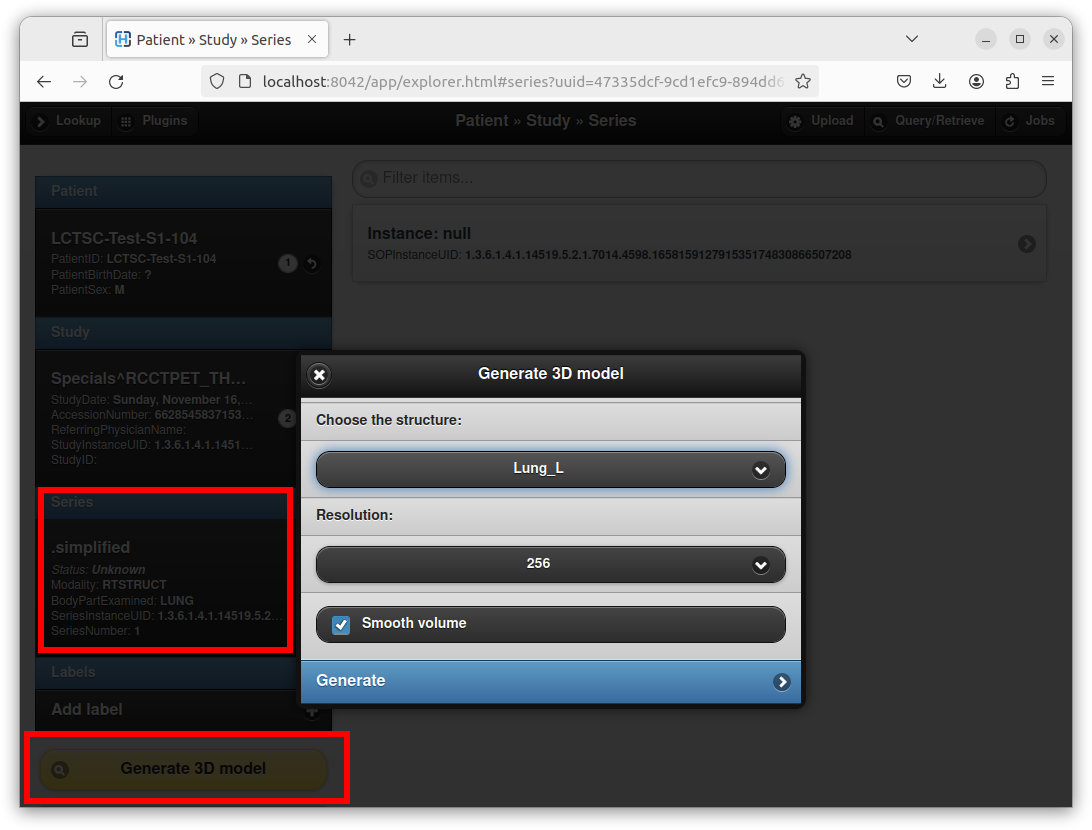
As can be seen in this screenshot, an interface opens to choose the structure set of interest, as well as the resolution of the intermediate 3D bitmap that will be used to create the STL mesh. After clicking on the “Generate” button, just like if attaching an existing STL file, Orthanc Explorer will open the newly created DICOM series and will propose to open a STL viewer.
Internally, the 3D model is generated using the well-known marching cubes algorithm, as implemented by the VTK library by Kitware. Additional technical details can be found in the reference paper.
Create STL from binary NIfTI¶
Besides converting RT-STRUCT to STL, it is also possible to convert a NIfTI 3D binary bitmap into a STL mesh. As this use case is very specific, it must be explicitly enabled in the configuration file of Orthanc as follows:
{
"Plugins" : [ "libOrthancSTL.so" ],
"STL" : {
"EnableNIfTI" : true
}
}
If the EnableNIfTI option is present, a new button entitled
“Attach NIfTI 3D model” appears if opening an existing DICOM study:
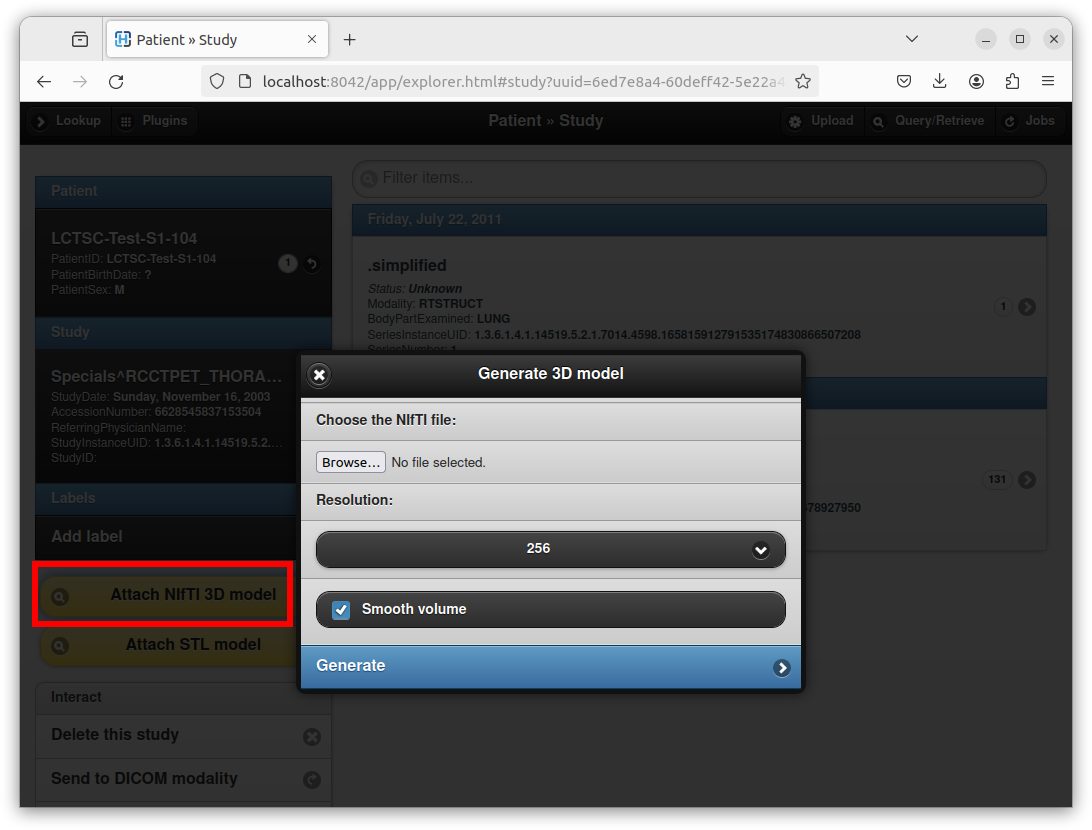
This dialog box can be used to upload a NIfTI volume, and to generate a 3D mesh with a specific resolution through the marching cubes algorithm.
REST API¶
Besides extending the Orthanc Explorer user interface, the STL plugin adds dedicated routes to the REST API of Orthanc.
DICOM-ization of STL files¶
The creation of a DICOM instance that embeds an existing STL file can
be done using the /tools/create-dicom route
in the built-in REST API of Orthanc (starting with version
1.12.1). This route can be used similarly to the DICOM-ization
of PDF files, with the data URI scheme using model/stl
instead of application/pdf. Here is a working example in Python 3:
import base64
import json
import requests
with open('liver.stl', 'rb') as f:
stl = f.read()
r = requests.post('http://localhost:8042/tools/create-dicom', json.dumps({
'Content' : 'data:model/stl;base64,%s' % base64.b64encode(stl).decode('ascii'),
'Parent' : '6ed7e8a4-60deff42-5e22a424-2128629f-158d0b3a',
'Tags' : {
'SeriesDescription' : 'Liver'
}
}))
r.raise_for_status()
instanceId = r.json() ['ID']
Note that if the Parent field is not provided, a new DICOM study
will be created.
Extraction of STL from DICOM¶
The route /instances/{id}/stl can be used to extract a STL from a
DICOM instance embedding a STL file, where id is the Orthanc
identifier of the DICOM instance. For instance:
$ curl http://localhost:8042/instances/a88c4c3f-8f2bd6fd-02080bed-92ab6817-2cb3c26e/stl > /tmp/liver.stl
$ meshlab /tmp/liver.stl
Evidently, an error is generated for DICOM instances that do not embed
a STL file. Note that meshlab is a well-known desktop application
to display STL file.
Listing structures of a DICOM RT-STRUCT¶
The STL plugin provides the list of the names of the structures that
are part of a DICOM RT-STRUCT instance with Orthanc identifier id at route /stl/rt-struct/{id}. For instance:
$ curl http://localhost:8042/stl/rt-struct/f0dc2345-8f627774-f66083ae-a14d781e-1187b513
[
"Esophagus",
"Heart",
"Lung_L",
"Lung_R",
"SpinalCord"
]
Generating a STL mesh from RT-STRUCT¶
A DICOM RT-STRUCT instance can be converted into a DICOM STL instance
using the /stl/encode-rtstruct route provided by the STL plugin.
Here is a sample Python script:
import json
import requests
r = requests.post('http://localhost:8042/stl/encode-rtstruct', json.dumps({
'Instance' : 'f0dc2345-8f627774-f66083ae-a14d781e-1187b513', # ID of the RT-STRUCT DICOM instance
'RoiNames' : [ 'Lung_L', 'Lung_R' ],
'Smooth' : True,
'Resolution' : 256
}))
r.raise_for_status()
instanceId = r.json() ['ID']
Note that contrarily to the default user interface, this route can be used to encode multiple structure sets as a single STL model.
Generating a STL mesh from NIfTI¶
Here is a sample Python 3 script to convert a NIfTI file as a DICOM STL instance:
import base64
import json
import requests
with open('colon.nii.gz', 'rb') as f:
nifti = f.read()
r = requests.post('http://localhost:8042/stl/encode-nifti', json.dumps({
'Nifti' : 'data:application/octet-stream;base64,' + base64.b64encode(nifti).decode('ascii'),
'ParentStudy' : '6ed7e8a4-60deff42-5e22a424-2128629f-158d0b3a',
'Smooth' : True,
'Resolution' : 256,
}))
r.raise_for_status()
instanceId = r.json() ['ID']